An Introduction to Database Building using NMRPredict DBA.
The key features of NMRPredict database building capability are:
- Databases can be built for 1H, 13C and X nuclei.
- Databases can be used by NMRServer and NMRPredict Desktop, and any 'in house' client program.
- Multiple structures can be added from an SDF file which includes assignments.
- Single entries can be added manually, by entering the assignments on the screen.
SDF file format.
The same format is used for 1H, 13C and X nuclei.
The SDF file format used for multiple entries is as follows, eg. for 13C.
MODGRAPH1234
3 2 0 0 0 0 0 0 0 0 2
3.9281 -3.0426 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9281 -1.7387 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6241 -3.4451 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
2 3 1 0 0 0 0
1 2 2 0 0 0 0
M END
> <ID>
9
> <SHIFT1>
1,133.39999,0.0
> <SHIFT2>
4,113.9,0.0
$$$$
where the first value is the atom number (in the MOL file) and the second number is the shift value.
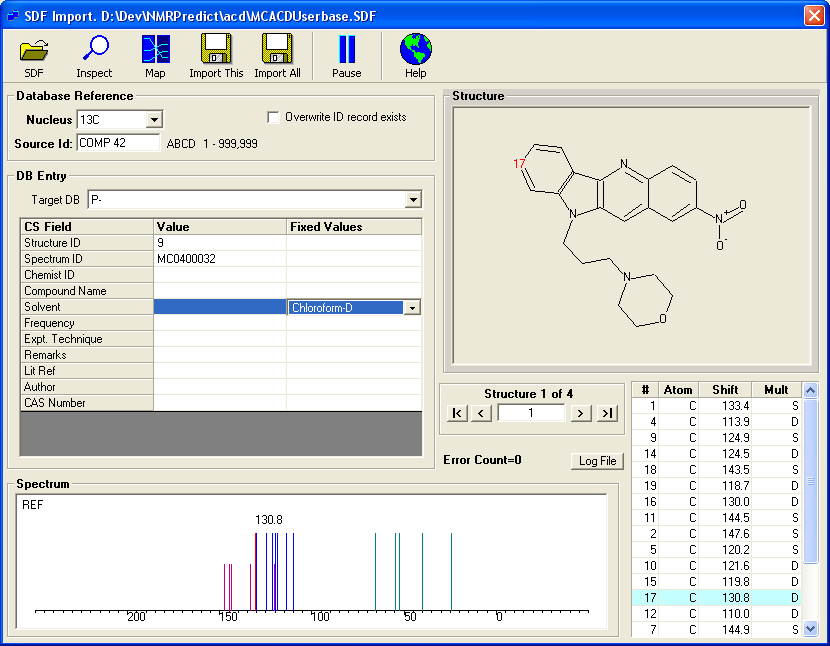
Loading an SDF file with 13C Assignments.
This screen shows the result of loading an SDF file with the above format.
- The application has read the first record and displays the spectrum, shifts and structure.
- Using 'tape buttons' you can scroll through the structures to examine the shifts.
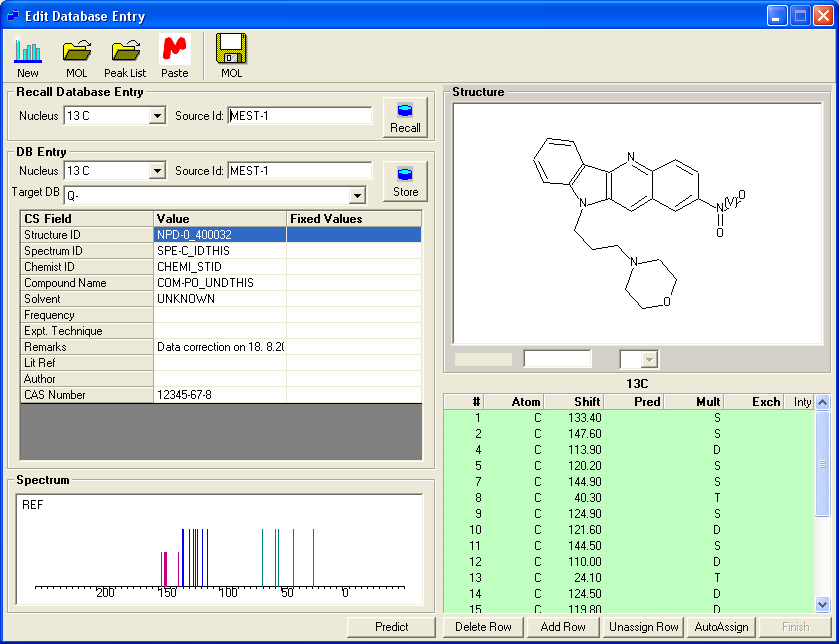
- By mapping the actual tag names in the SDF file the application is able to read items such solvent, chemist and id. The database fields, 'Structure Id', 'Spectrum Id' and 'Chemist Id' can be used to search the database.
- The user has to choose a database letter 'N' to 'Z'. The database letter chosen rpresents the unit which can be easily be excluded from any subsequent prediction or search. It may also be deleted and recreated. Thus, the database letter should be carefully selected on a organisation wide basis.
- The 'Source Id' must also be selected. This consists of four letters and a number between 1 and 99,999. This can further be used to group structures. This also allows groups of structures to be allocated a project or departmental code.
Assigning13C Shifts in a MOL file.
To assign the shifts on a single structure, load the MOL file, click on an atom; the cursor will then automatically move to the shift text box, when the shift value can be typed. The structure can then be predicted to check the assigned values.
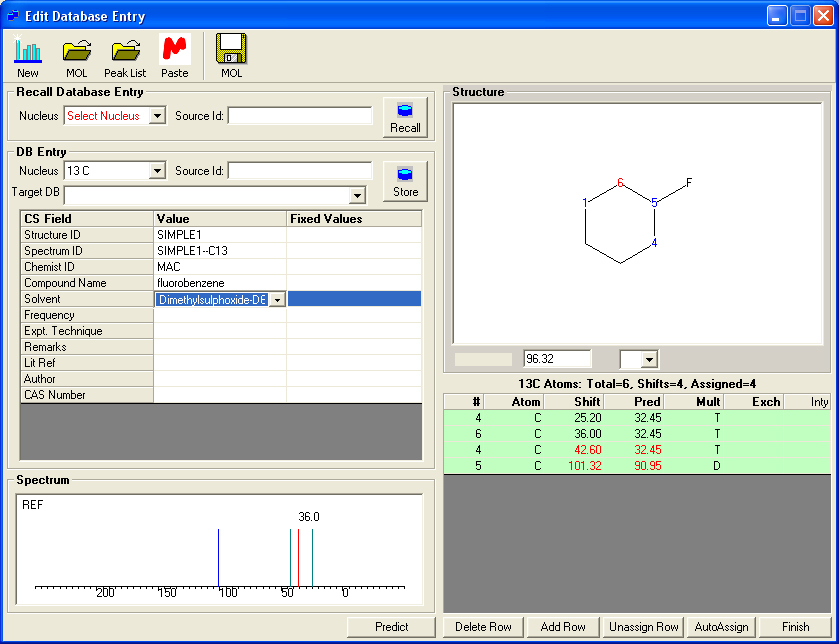
Editing a 13C Database Entry.
To edit an existing entry in a 13C or X nuclei database recall the entry by selecting the nucleus and 'Source Id'. The shift values may then be editted or new shift values assigned.
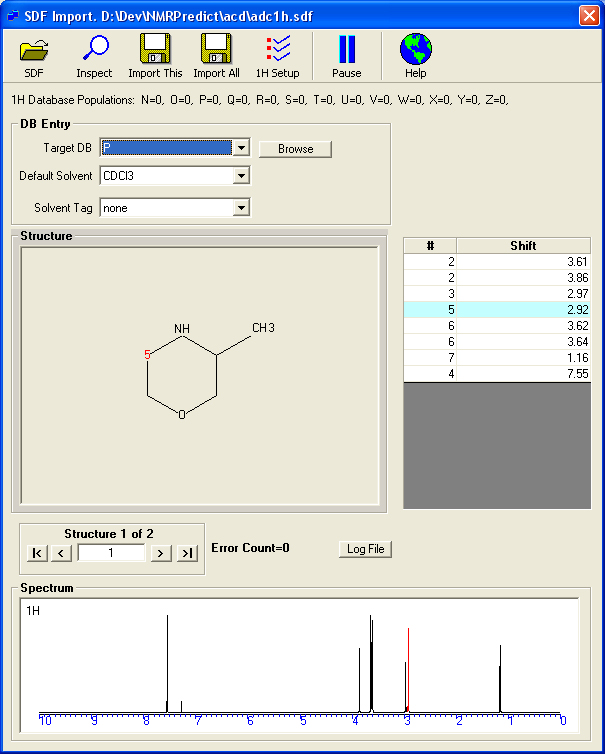
Loading an SDF file with 1H Assignments.
The procedure for a 1H datagbase is the same as 13C.
- Loading the SDF file with the above format.
- The application will read the first record and display the spectrum, shifts and structure.
- Using 'tape buttons' you can scroll through the structures to examine the shifts.
- By mapping the actual tag names in the SDF file the application is able to read the solvent for each record.
- The user has to choose a database letter 'N' to 'Z'. The database letter chosen rpresents the unit which can be easily be excluded from any subsequent prediction or search. It may also be deleted and recreated. Thus the database letter should be carefully selected on a organisation wide basis.
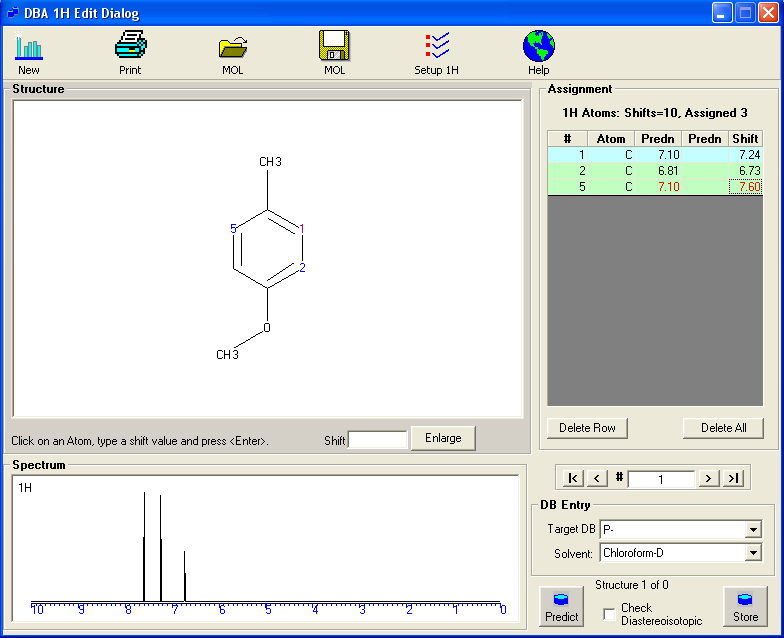
Assigning 1H Shifts in a MOL file.
To assign the shifts on a single structure, load the MOL file, click on an atom; the cursor will then automatically move to the shift text box, when the shift value can be typed. The spectrum is displayed as the atoms are assigned. The structure can then be predicted to check the assigned values..